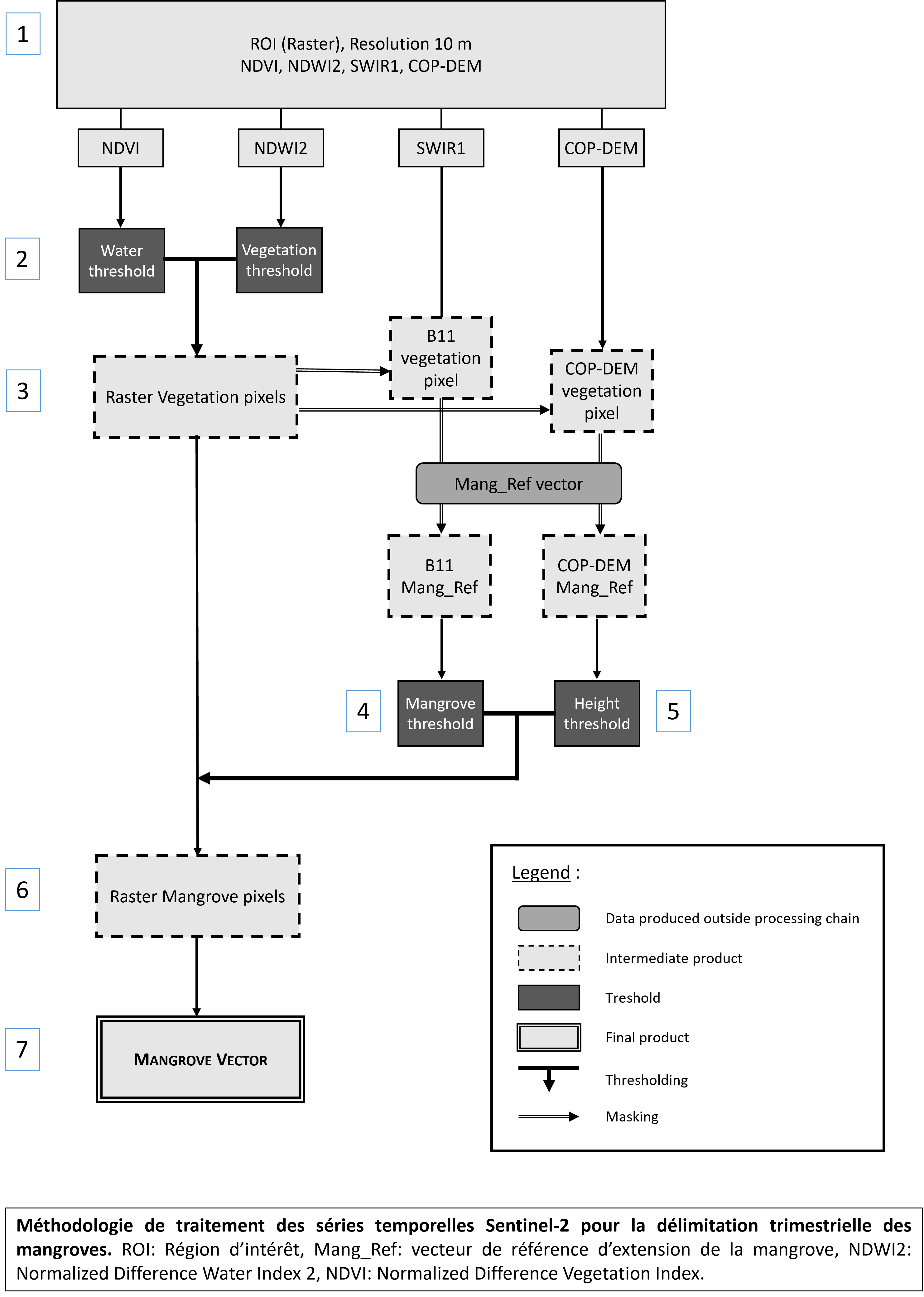
A processing chain for delimiting mangrove surfaces has been developed. It is iterative: for each image, at each quarter, a mangrove contour vector is calculated from the least cloudy Sentinel L2A image of the quarter. The mangrove vector can be viewed and downloaded in SHP format free of charge.
For a given quarter and a given tile, the input data are :
- A mangrove extension reference vector (Mang_Ref). The Mang_ref reference vector is a mangrove contour vector from the scientific community
- A vector of the region of interest (ROI) in which the algorithm will search for mangrove pixels. The ROI is defined from the Mang_Ref vector to which an external buffer of 500m is applied
- The SWIR1 band (B11) of the Sentinel-2 L2A image, the least cloudy of the quarter, with a spatial resolution of 20m
- Two criteria are used to select the least cloudy image:
1) a time lag of at least 30 days with the date of the image selected for the previous quarter
2) a minimum rate of clouding in the area corresponding to mang-Ref
- The Copernicus DEM (Global Digital Elevation Model - COP-DEM, https://doi.org/10.5270/ESA-c5d3d65), spatial resolution 30m
- The NDWI2 raster, spatial resolution 10m
- NDVI raster, spatial resolution 10m

The following processes are applied :
- Creation of the raster in which the search for mangrove pixels is carried out
A 4-band raster (NDWI2, NDVI, SWIR1 and COP-DEM) with 10m resolution is created: Raster_M.
The Raster_M is cut with the ROI vector. - Definition of “land” and “vegetation” thresholds with the NDWI2 and NDVI indices
The NDWI2 (Normalized Difference Water Index 2) is used to detect flooded surfaces, and ranges from -1 to 1. Pixels with an NDWI2 value below a threshold S1 (for example 0) define land surfaces. The threshold can be adjusted by the administrator depending on the study area. The NDVI (Normalized Vegetation Difference Index) is used to detect vegetated surfaces. It ranges between -1 and 1, with pixels with an NDVI value greater than a threshold S2 (for example 0,3) defining vegetated surfaces. The threshold can be adjusted by the administrator depending on the study area. - Selecting vegetation pixels.
Double thresholding is appliedon SWIR1 and COP-DEM bands. Vegetation pixels have an NDWI2 value < S1 AND an NDVI value > S2. - Define the values of the mangrove pixels in the SWIR1 band.
From the SWIR1 band and in the geographical extension of Mang_Ref, two thresholds are extracted: the 1% quantile and the 98% quantile. These thresholds will define in which value range the mangroves are located. The thresholds can be adjusted according to the study zones by the administrator. - Define the values of the mangrove pixels in the COP-DEM band.
From the COP-DEM band and within the geographical extension of Mang_Ref, the maximum mangrove height threshold (Hmax) is extracted. - Selection of mangrove pixels using the SWIR1 band and the COP-DEM band.
Double thresholding is applied. Mangrove pixels have a COP-DEM value ≤ Hmax AND a value of Q1 < SWIR1 < Q98. - Vectorisation of the mangrove raster.
Creation of a vector shapefile of mangrove contours.
